We are happy to announce that JIT support in both the PowerPC backend and the
s390x backend have been enhanced. Both can now vectorize loops via SIMD
instructions. Special thanks to IBM for funding this work.
If you are not familiar with this topic you can read more details here.
There are many more enhancements under the hood. Most notably, all pure operations are now delayed until the latest possible point. In some cases indices have been calculated more than once or they needed an additional register, because the old value is still used. Additionally it is now possible to load quadword-aligned memory in both PPC and s390x (x86 currently cannot do that).
The work done here to support vectorization helps micronumpy (NumPyPy) to speed up operations for PPC and s390x. So why is PyPy supporting both NumPyPy and NumPy, do we actually need both? Yes, there are places where gcc can beat the JIT, and places where the tight integration between NumPyPy and PyPy is more performant. We do have plans to integrate both, hijacking the C-extension method calls to use NumPyPy where we know NumPyPy can be faster.
Just to give you an idea why this is a benefit:
NumPy arrays can carry custom dtypes and apply user defined python functions on the arrays. How could one optimize this kind of scenario? In a traditional setup, you cannot. But as soon as NumPyPy is turned on, you can suddenly JIT compile this code and vectorize it.
Another example is element access that occurs frequently, or any other calls that cross between Python and the C level frequently.
functioning properly. Additionally it has been rewritten to use perf instead of the timeit stdlib module.
x86 ran on Fedora 24 (kernel version of 4.8.4), PPC ran on Fedora 21 (kernel version 3.17.4) and s390x ran on Redhat Linux 7.2 (kernel version 3.10.0). Respectivley, numpy on cpython had openblas available on x86, no blas implementation were present on s390x and PPC provided blas and lapack.
As you can see all machines run very different configurations. It does not make sense to compare across platforms, but rather implementations on the same platform.
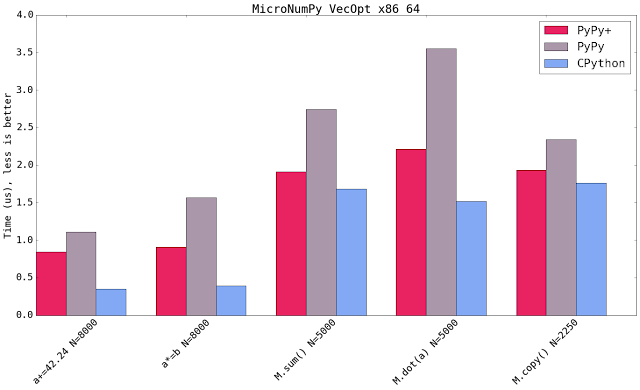
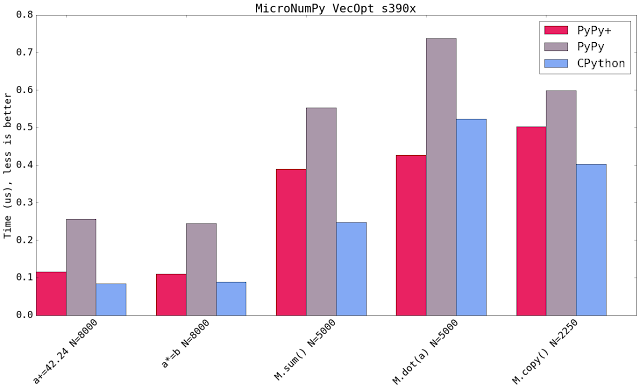
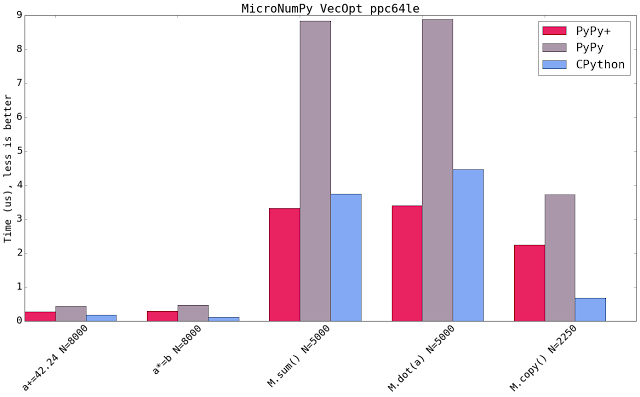
Blue shows CPython 2.7.10+ available on that platform using the latest NumPy (1.11). Micro NumPy is used for PyPy. PyPy+ indicates that the vectorization optimization is turned on.
All bar charts show the median value of all runs (5 samples, 100 loops, 10 inner loops, for the operations on vectors (not matrices) the loops are set to 1000). PyPy additionally gets 3 extra executions to warmup the JIT.
The comparison is really comparing speed of machine code. It compares the PyPy's JIT output vs GCC's output. It has little to do with the speed of the interpreter.
Both new SIMD backends speedup the numeric kernels. Some times it is near to the speed of CPython, some times it is faster. The maximum parallelism very much depends on the extension emitted by the compiler. All three SIMD backends have the same vector register size (which is 128 bit). This means that all three behave similar but ppc and s390x gain more because they can load 128bit of memory from quadword aligned memory.
PyPy can make a valuable contribution for data scientists, helping them to rapidly write scientific programs in Python and run them at near native speed. If you happen to be in that situation, we are eager to hear you feedback or resolve your issues and also work together to improve the performance of your,
code. Just get in touch!
Richard Plangger (plan_rich) and the PyPy team
s390x backend have been enhanced. Both can now vectorize loops via SIMD
instructions. Special thanks to IBM for funding this work.
If you are not familiar with this topic you can read more details here.
There are many more enhancements under the hood. Most notably, all pure operations are now delayed until the latest possible point. In some cases indices have been calculated more than once or they needed an additional register, because the old value is still used. Additionally it is now possible to load quadword-aligned memory in both PPC and s390x (x86 currently cannot do that).
NumPy & CPyExt
The community and core developers have been moving CPyExt towards a complete, but emulated, layer for CPython C extensions. This is great, because the one restriction preventing the wider deployment of PyPy in several scenarios will hopefully be removed. However, we advocate not to use CPyExt, but rather to not write C code at all (let PyPy speed up your Python code) or use cffi.The work done here to support vectorization helps micronumpy (NumPyPy) to speed up operations for PPC and s390x. So why is PyPy supporting both NumPyPy and NumPy, do we actually need both? Yes, there are places where gcc can beat the JIT, and places where the tight integration between NumPyPy and PyPy is more performant. We do have plans to integrate both, hijacking the C-extension method calls to use NumPyPy where we know NumPyPy can be faster.
Just to give you an idea why this is a benefit:
NumPy arrays can carry custom dtypes and apply user defined python functions on the arrays. How could one optimize this kind of scenario? In a traditional setup, you cannot. But as soon as NumPyPy is turned on, you can suddenly JIT compile this code and vectorize it.
Another example is element access that occurs frequently, or any other calls that cross between Python and the C level frequently.
Benchmarks
Let's have a look at some benchmarks reusing mikefc's numpy benchmark suite (find the forked version here). I only ran a subset of microbenchmarks, showing that the core functionality isfunctioning properly. Additionally it has been rewritten to use perf instead of the timeit stdlib module.
Setup
x86 runs on a Intel i7-2600 clocked at 3.40GHz using 4 cores. PowerPC runs on the Power 8 clocked at 3.425GHz providing 160 cores. Last but not least the mainframe machine clocked up to 4 GHz, but fully virtualized (as it is common for such machines). Note that PowerPC is a non private remote machine. It is used by many users and it is crowded with processes. It is hard to extract a stable benchmark there.x86 ran on Fedora 24 (kernel version of 4.8.4), PPC ran on Fedora 21 (kernel version 3.17.4) and s390x ran on Redhat Linux 7.2 (kernel version 3.10.0). Respectivley, numpy on cpython had openblas available on x86, no blas implementation were present on s390x and PPC provided blas and lapack.
As you can see all machines run very different configurations. It does not make sense to compare across platforms, but rather implementations on the same platform.



Blue shows CPython 2.7.10+ available on that platform using the latest NumPy (1.11). Micro NumPy is used for PyPy. PyPy+ indicates that the vectorization optimization is turned on.
All bar charts show the median value of all runs (5 samples, 100 loops, 10 inner loops, for the operations on vectors (not matrices) the loops are set to 1000). PyPy additionally gets 3 extra executions to warmup the JIT.
The comparison is really comparing speed of machine code. It compares the PyPy's JIT output vs GCC's output. It has little to do with the speed of the interpreter.
Both new SIMD backends speedup the numeric kernels. Some times it is near to the speed of CPython, some times it is faster. The maximum parallelism very much depends on the extension emitted by the compiler. All three SIMD backends have the same vector register size (which is 128 bit). This means that all three behave similar but ppc and s390x gain more because they can load 128bit of memory from quadword aligned memory.
Future directions
Python is achieving rapid adoption in data science. This is currently a trend emerging in Europe, and Python is already heavily used for data science in the USA many other places around the world.PyPy can make a valuable contribution for data scientists, helping them to rapidly write scientific programs in Python and run them at near native speed. If you happen to be in that situation, we are eager to hear you feedback or resolve your issues and also work together to improve the performance of your,
code. Just get in touch!
Richard Plangger (plan_rich) and the PyPy team
4 comments:
As you are talking about GCC beating your JIT, you are using your own vectorizing compiler right?
I wonder if this is a feasible approach. Can you really compete with the years if not decades of work that went into the vectorizers of GCC and LLVM?
Wouldn't it make more sense to plug into GCC's and LLVM's JIT API's (yes GCC has a JIT) for this type of code?
What does PyPy bring to the table that the existing JIT's do not for numerical code?
It's good to see pypy making progress on using python as a toolkit for data science. In addition to numpy, pandas/scipy also needs to work well for me to switch.
Also, a lot of data science is currently being run on windows and the x64 port of pypy hasn't had much traction in the last several years. If these 2 issues are solved (pandas/scipy being supported on a x64 windows pypy) then there should be no reason to keep using CPython.
I think most of pypy dev/users use Linux/MacOsx only, so there is no strong motivation to support win64 at the moment.
Not necessarily the users---there are some on Windows. But the point is that we have not a single developer on Windows. Until someone comes forward with a serious offer for either code or money, Win64 will not get magically done.
Post a Comment